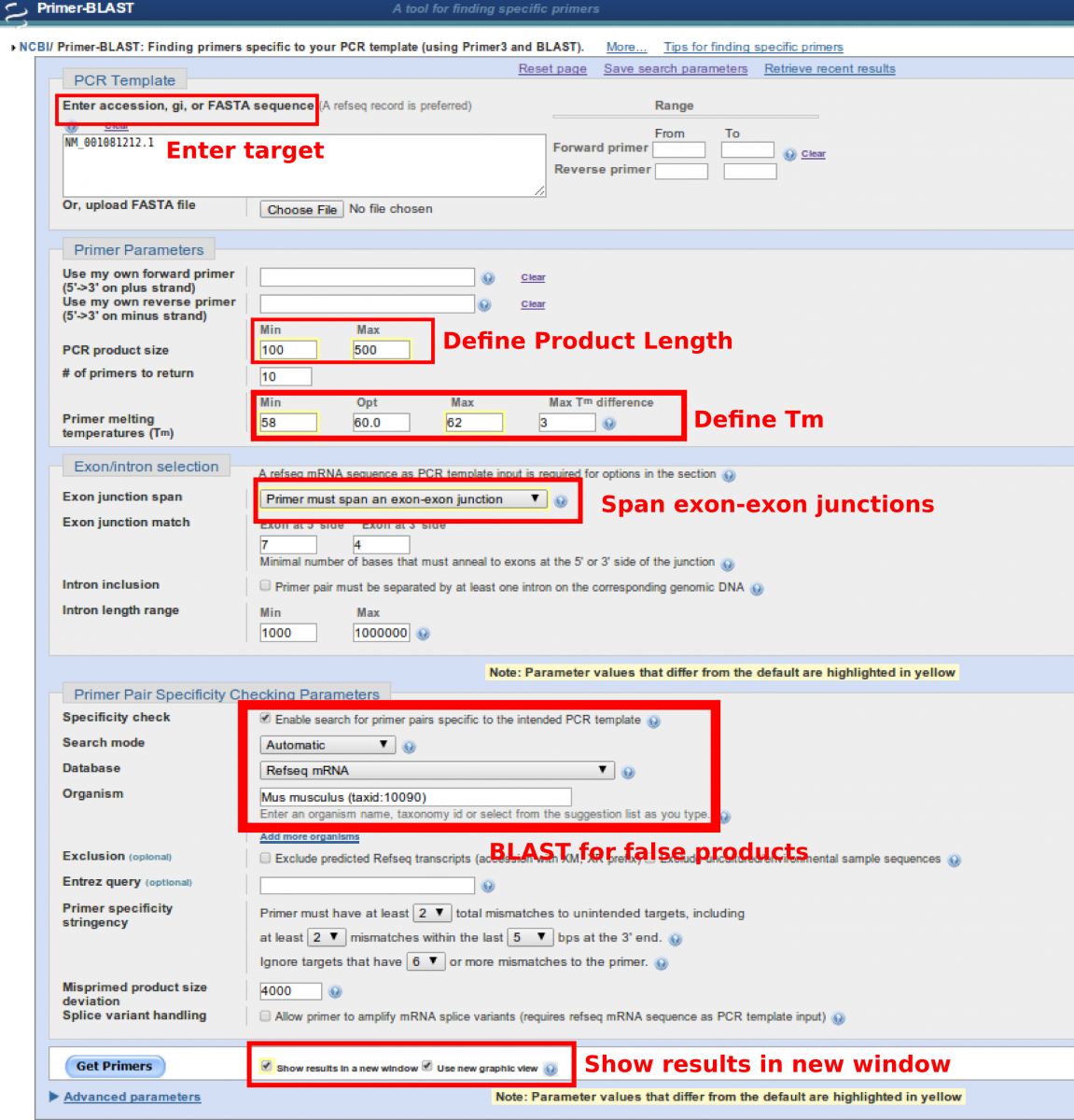
Primer-BLAST is a combination of a program called Primer3 that aids in the design of primers with specific properties and BLAST. Primer-BLAST allows for the construction of primers for qPCR where the user can specify the melting temperature, reduce the amount of self-priming and to span exon-exon junctions in order to avoid amplification of contaminating genomic DNA. After the design of primers, each primer pair is sent into BLAST to identify if similar products within the genome of the model organism will also be primed and amplified. This process ensures that the primers designed fall within your design parameters and most likely only amplify your gene of interest.
- Enter the sequence OR the NCBI accession number for the gene of interest
- Define the PCR product length
- limiting the product between 100-500 permits for good efficiency in thqPCR
- longer products may not be efficiently replicated depending on your cycling protocol
- Define the desired melting temperature (Tm) of the primers (minimum, optimal, maximum, difference between the set)
- 60ºC is fairly high and will aid in the enhanced specificity of the primer with the target during amplification to avoid false priming
- Try to have the Tm as close as possible so that they are annealing about equally
- Choose the option “Primer must span an exon-exon junction”
- This aids in amplifying cDNA and not genomic DNA that may be contaminating
- do not select this if it a single exon gene as this will fail
- Select Refseq mRNA as the database to search against.
- Refseq provides sequences to naturally occurring sequences.
- Things like plasmid sequences or vector constructs do not show up in Refseq
- Select the organism you are BLASTing against
- there are options for model organisms as well as cell lines
- If you are using something like PC12 cells, you may use Rattus norvegicus or PC12 genome since that is also an option in the database
- Evaluate the location of the primers and the other parameters. We generally choose primers at the 3′ end of the RNA since RT reactions often have a 3′ bias in eukaryotes by using oligo-dT priming in the reverse transcription