RNA-Seq
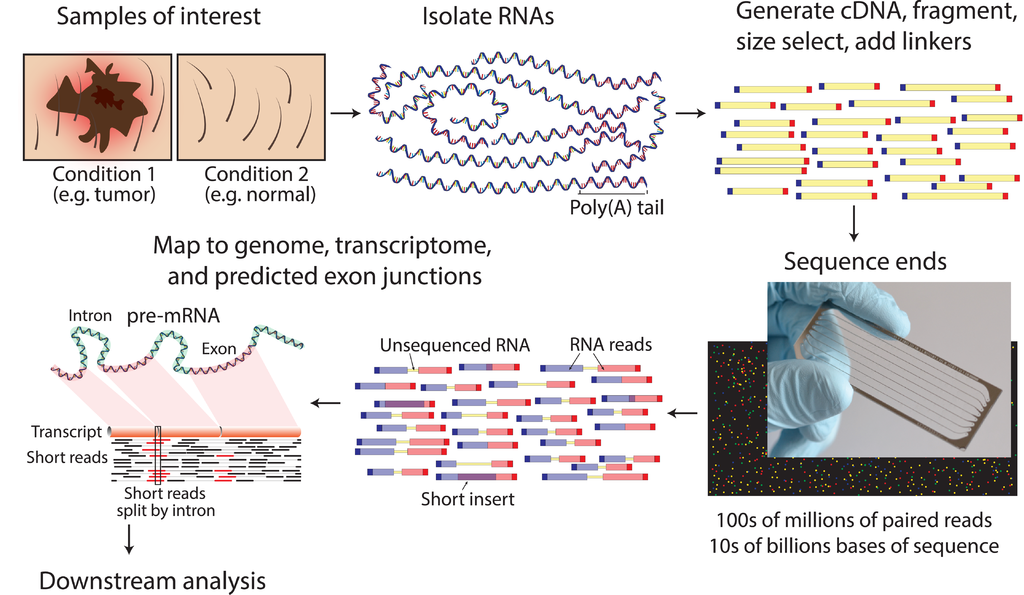

Given sufficient read coverage, novel splice isoforms can also be identified as different exon-exon junctions are identified.
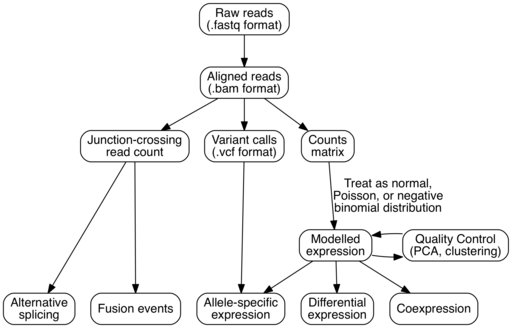
The general workflow of RNA-Seq analysis follows:
Molecular & Cell Biology Lecture
A Reading supplement for Molecular and Cell Biology


Given sufficient read coverage, novel splice isoforms can also be identified as different exon-exon junctions are identified.
The general workflow of RNA-Seq analysis follows:
