Restriction Enzymes
DNA can be cut by restriction endonucleases (RE). Endonucleases are enzymes that can hydrolyze the nucleic acid polymer by breaking the phosphodiester bond between the phosphate and the pentose on the nucleic acid backbone. This is a very strong covalent bond while the weaker hydrogen bonds maintain their interactions and double strandedness.
As the name implies, restriction endonucleases (or restriction enzymes) are “restricted” in their ability to cut or digest DNA. The restriction that is useful to biologists is usually palindromic DNA sequences. Palindromic sequences are the same sequence forwards and backwards. Some examples of palindromes: RACE CAR, CIVIC, A MAN A PLAN A CANAL PANAMA. With respect to DNA, there are 2 strands that run antiparallelel to each other. Therefore, the reverse complement of one strand is identical to the other. Molecular biologists also tend to use these special molecular scissors that recognize palindromes of 6 or 8. By using 6-cutters or 8-cutters, the sequences occur throughout large stretches rarely, but often enough to be of utility.



Restriction enzymes hydrolyze covalent phosphodiester bonds of the DNA to leave either “sticky/cohesive” ends or “blunt” ends. This distinction in cutting is important because an EcoRI sticky end can be used to match up a piece of DNA cut with the same enzyme in order to glue or ligate them back together. While endonucleases cut DNA, ligases join them back together. DNA digested with EcoRI can be ligated back together with another piece of DNA digested with EcoRI, but not to a piece digested with SmaI. Another blunt cutter is EcoRV with a recognition sequence of GAT | ATC.
Features of Plasmids
Plasmids that are designed by Biologists to shuttle pieces of DNA for study are referred to as vectors, because they move a piece of DNA.
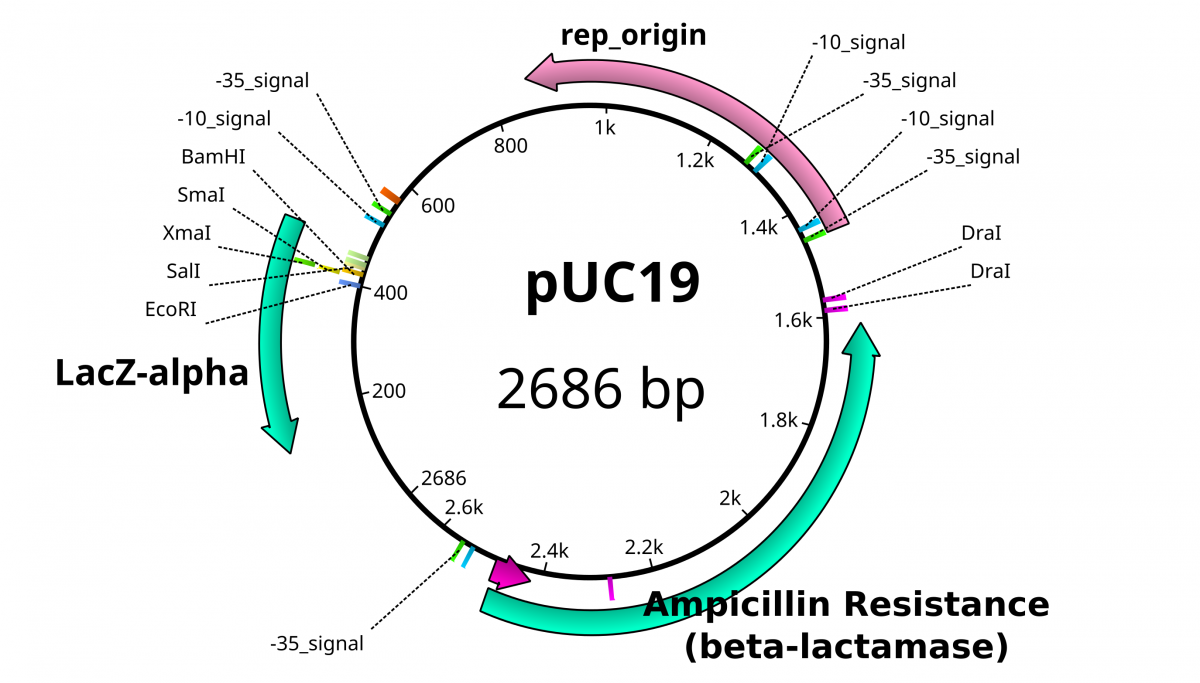 These plasmid vectors have the same hallmarks as traditional plasmids with the capacity to replicate independently of the bacterial genome. The feature that allows these DNA’s to replicate is called an origin of replication (ori) that is usually rich in A’s and T’s. However, these plasmid vectors have the additional properties that make them easy to work with and distinguishable from bacterial plasmids; a selection marker and a multiple cloning site. A selection marker usually comes in the form of a gene that encodes resistance to a specific antibiotic. In the pictured plasmid, Ampicillin resistance granted by the β-lactamase gene. The multiple cloning site (MCS), also known as the polylinker, is the location in which the DNA of interest is incorporated into the vector. MCSs are defined by a set of unique sites where the DNA can be cut by restriction endonucleases (RE). As the name implies, restriction enzymes are “restricted” in their ability to cut or digest DNA. The restriction that is useful to biologists is usually palindromic DNA sequences.With respect to DNA, there are 2 strands that run antiparallelel to each other. Therefore, the reverse complement of one strand is identical to the other.
These plasmid vectors have the same hallmarks as traditional plasmids with the capacity to replicate independently of the bacterial genome. The feature that allows these DNA’s to replicate is called an origin of replication (ori) that is usually rich in A’s and T’s. However, these plasmid vectors have the additional properties that make them easy to work with and distinguishable from bacterial plasmids; a selection marker and a multiple cloning site. A selection marker usually comes in the form of a gene that encodes resistance to a specific antibiotic. In the pictured plasmid, Ampicillin resistance granted by the β-lactamase gene. The multiple cloning site (MCS), also known as the polylinker, is the location in which the DNA of interest is incorporated into the vector. MCSs are defined by a set of unique sites where the DNA can be cut by restriction endonucleases (RE). As the name implies, restriction enzymes are “restricted” in their ability to cut or digest DNA. The restriction that is useful to biologists is usually palindromic DNA sequences.With respect to DNA, there are 2 strands that run antiparallelel to each other. Therefore, the reverse complement of one strand is identical to the other.


Restriction enzymes hydrolyze covalent phosphodiester bonds of the DNA to leave either “sticky/cohesive” ends or “blunt” ends. This distinction in cutting is important because an EcoRI sticky end can be used to match up a piece of DNA cut with the same enzyme in order to glue or ligate them back together. While endonucleases cut DNA, ligases join them back together. DNA digested with EcoRI can be ligated back together with another piece of DNA digested with EcoRI, but not to a piece digested with SmaI. Another blunt cutter is EcoRV with a recognition sequence of GAT | ATC.
By “cutting and pasting” DNA into vectors, we can introduce foreign or exogenous DNA into bacteria. This type of DNA is now called Recombinant DNA and is the heart of biotechnology.