Contents
Proteins are polymers of Amino Acids
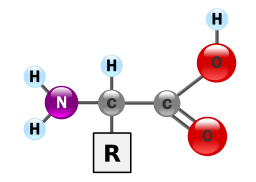 Proteins provide much of the structural and functional capacity of cells. Proteins are composed of monomers called amino acids. Amino Acids are hydrocarbons that have an amino group (-NH2) and an acidic carboxyl group (-COOH).The R group represents a hydrocarbon chain with a modification that alters the properties of the amino acid. 20 universal amino acids are used to construct proteins. The variation in functional groups along the amino acid chain gives rise to the functional diversity of proteins.
Proteins provide much of the structural and functional capacity of cells. Proteins are composed of monomers called amino acids. Amino Acids are hydrocarbons that have an amino group (-NH2) and an acidic carboxyl group (-COOH).The R group represents a hydrocarbon chain with a modification that alters the properties of the amino acid. 20 universal amino acids are used to construct proteins. The variation in functional groups along the amino acid chain gives rise to the functional diversity of proteins.


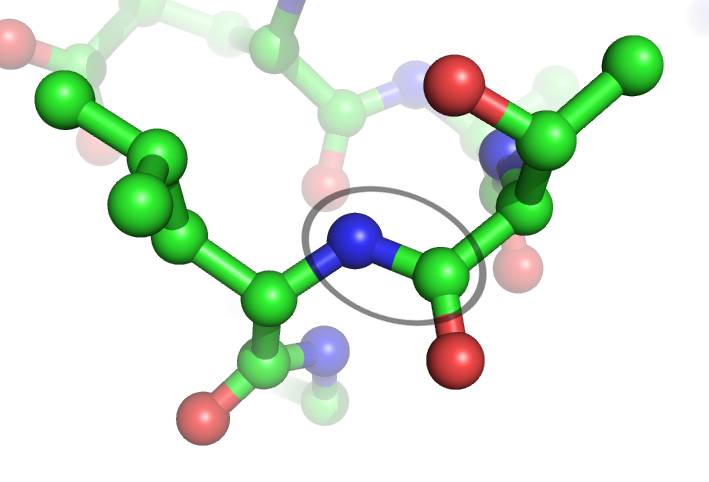
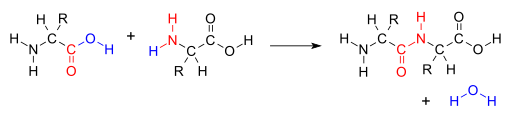
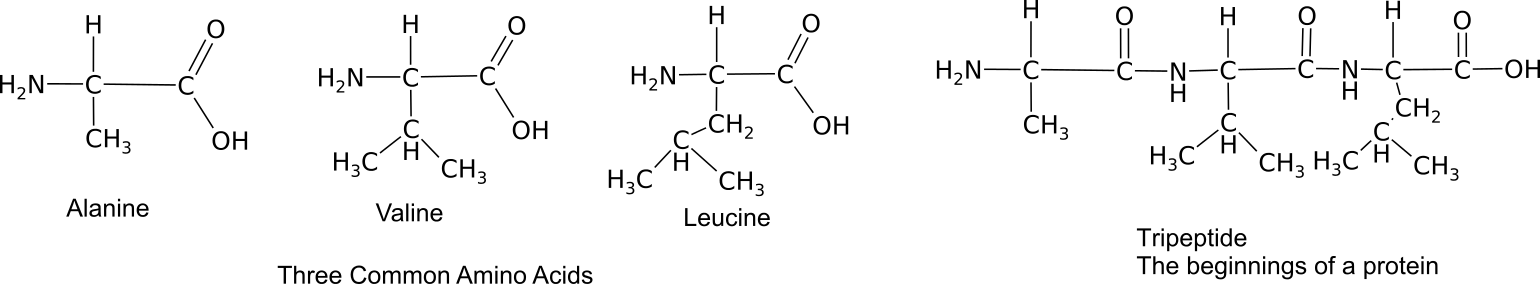
How amino acids interact with each other and the environment
Use the following simulation to test how a polypeptide chain with fold based on the type of solution it is in and the composition of the amino acids.
- Protein Folding Simulation (CC BY 4.0 Concord Consortium)
Levels of structure
Primary Structure (1°)
The sequence of amino acids read from the Amino or N-terminal end of the molecule to the Carboxyl or C-terminal end
Tyr-Cys-Arg-Phe-Leu-Val-….
Secondary Structure (2°)
local three-dimensional structures that form from interactions of amino acids, like hydrogen bonding
Alpha Helix
coils occurring from the H-bonds between N-H and C=O groups along the backbone of the protein
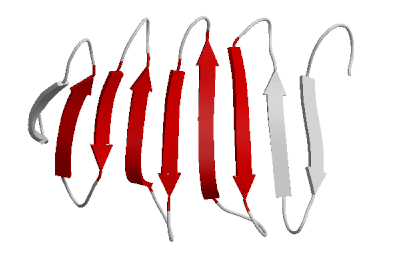
Beta Sheets
laterally connected strands or sheets of amino acids occurring from the H-bonds between N-H and C=O groups along the backbone of the protein
-
-
Secondary Structure Proclivity
The repetitive nature of the peptide bond creates a largely planar three-dimensional structure. Torsion around the bonds offer some flexibility and the sidechain properties yield properties that define the degree of torsion. The Ramachandran Plot or [φ,ψ plot] was developed in the 1960s to model the energetically favorable states of these dihedral angles. The dihedral angle between an N and Cα is called φ while the dihedral angle between an Cα and Cβ is called ψ.

By modeling the angle propensities, of an amino acid sequence, the likelihood of α-helices and β-sheets can be predicted.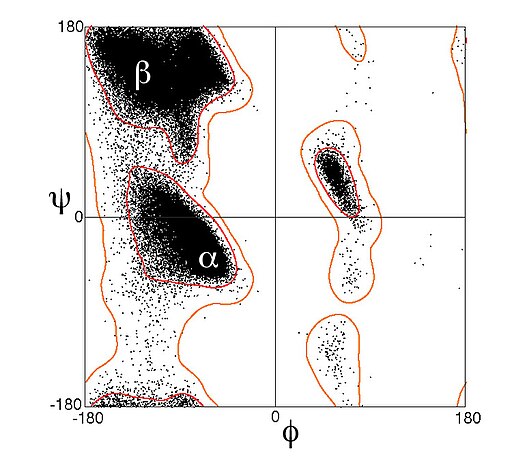
Ramachandran plot of data from Lovell et al. 2003 , showing about 100,000 data points for general amino-acid types (not Gly, Pro, or pre-Pro) in high-resolution crystal structures. Tertiary structure (3°)
The overall 3-D structure of the polypeptide chain is influenced by many intramolecular interactions and altered by interactions with external molecules.

Quaternary structure(4°)
Some proteins are composed of multiple polypetide chains and are multimeric.
An example of a multimeric protein is an IgG antibody.

Another example of a multimeric protein with quaternary structure is the hemoglobin molecule composed of 4 globin proteins.
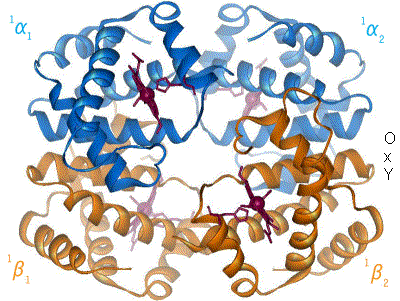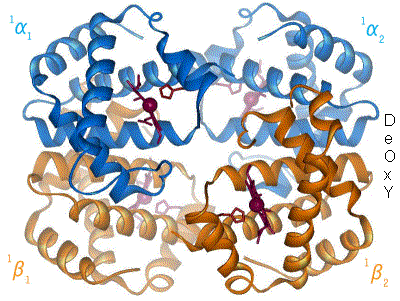
Diversity of Proteins
Interactive of Protein Diversity at the Protein Data Bank
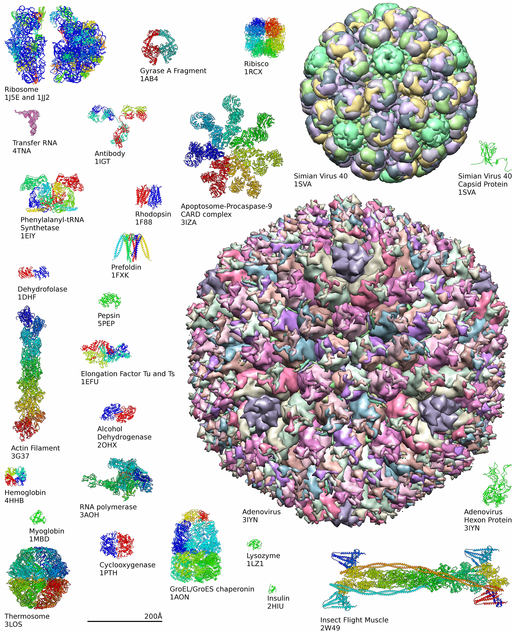
Learn more about complexity of protein structures at the Protein Data Bank
-