Expansion of sequencing technology

Traditional sequencing of genomes was a long and tedious process that cloned fragments of genomic DNA into plasmids to generate a genomic DNA library (gDNA). These plasmids were individually sequenced using Sanger sequencing methodology and computational was performed to identify overlapping pieces, like a jigsaw puzzle. This assembly would result in a draft scaffold.
As technology improved, the cost of sequencing genomes became less expensive. This technology outpaced the Moore’s Law, a semiconductor projection about the the speed of computers as time progressed. A dramatic price decrease in cost of genome sequencing occurred around 2008 due to technical advances.
 As the cost of genome sequencing decreased, a dramatic increase in genome deposition into Genbank was observed. These deposits reflected small genomes of bacteria and archaea.
As the cost of genome sequencing decreased, a dramatic increase in genome deposition into Genbank was observed. These deposits reflected small genomes of bacteria and archaea.

Credit: Estevezj (CC-BY-SA 3.0)
The decrease in per nucleotide sequencing cost came from the parallelization of sequencing. Whereas Sanger Sequencing is capable of sequencing one stretch at a time, a parallel assembly of sequencing reactions has lead to high throughput sequencing often dubbed Next Generation Sequencing (NGS).
The Next Generation of Sequencing: High-Throughput Technologies
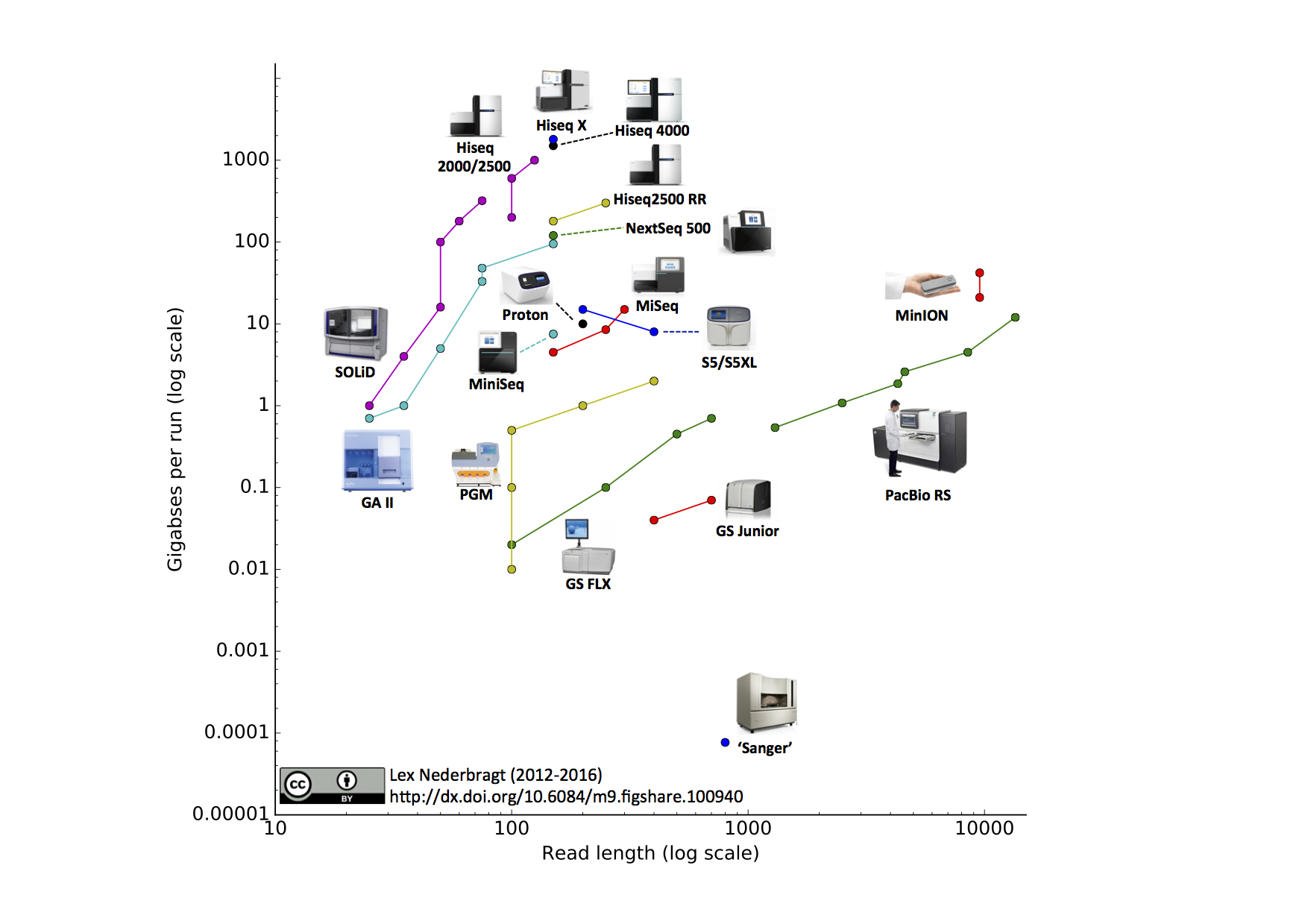
High Throughput Sequencing Applied to Genome Sequencing (TEDed CC BY-NC-ND 4.0)
Short Read Sequencing by Synthesis
Illumina
Illumina short read sequencing uses flow cell technology where oligonucleotides complimentary to adapter primers are physically seeded.



Ion Torrent
Fragmented DNA is ligated to adapter sequences and adhered onto microbeads. The beads are embedded into microwells on a semiconductor. Ion Torrent performs the sequencing reactions in an unbuffered solution since the semiconductor acts as a pH meter to identify nucleotide incorporation. Standard nucleotides are flooded onto the chip and incorporated. Because nucleotide incorporation creates a proton (H+), a microenvironment of low pH is detected in the unbuffered solution.
Single Molecule Real Time Sequencing
Pac Bio
Pac Bio uses nanowells with covalent bonded DNA polymerase to sequence individual molecules of DNA. Fluorescent nucleotides are incorporated during synthesis reactions and a real-time incorporation can be measured. Pac Bio sequencing has the advantage of sequencing fragments of 10-20kb, in stark contrast to the short read methods.
Oxford Nanopore
Credit: George Church (CC-BY 3.0)
Oxford Nanopore utilizes the protein alpha-hemolysin integrated onto a semiconductor chip. The pore size of the protein is the correct ssize for a single DNA molecule to fit through. A DNA Polymerase molecule is linked to the opening of the pore where the replicated DNA is fed through. As the DNA traverses the pore, the voltage changes are measured and mapped to the qualities of specific bases.
https://youtu.be/BNz880V52rQ