Table of Contents
Polymers of Amino Acids
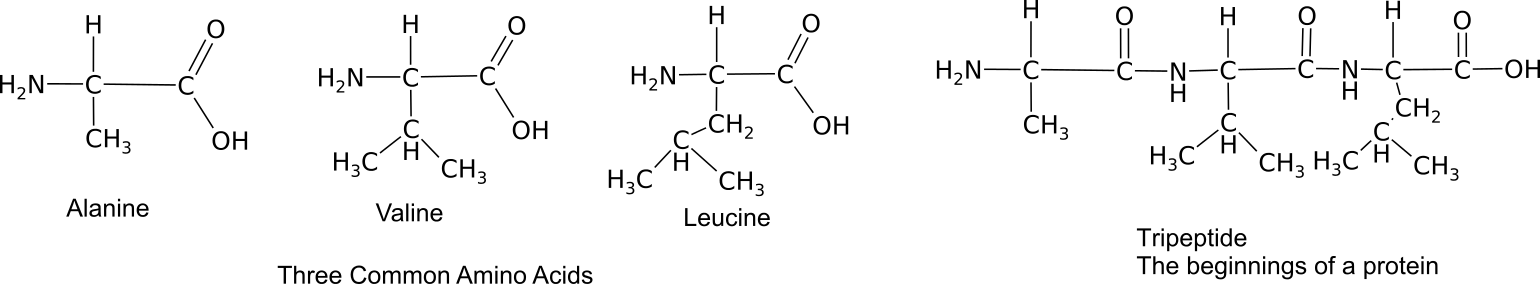
Proteins provide much of the structural and functional capacity of cells. Proteins are composed of monomers called amino acids. Amino acids are hydrocarbons that have an amino group (-NH2) and an acidic carboxyl group (-COOH).The R group represents a hydrocarbon chain with a modification that alters the properties of the amino acid. 20 universal amino acids are used to construct proteins. The variation in functional groups along the amino acid chain gives rise to the functional diversity of proteins.
Monomers bond together through a dehydration synthesis reaction between adjacent amino and carboxyl groups to yield a peptide bond.
How Amino Acids Interact with Their Environment
Use the following simulation to test how a polypeptide chain with fold based on the type of solution it is in and the composition of the amino acids. You can use the buttons on the bottom of the simulation to toggle between hydrophilic amino acids and hydrophobic amino acids. You can also change the background solution. Make note of how the different types of amino acids react in water and in oil.
Protein Folding Simulation (CC BY 4.0 Concord Consortium)
Levels of Protein Structure
Primary structure (1°) is the sequence of amino acids in the protein. This sequence is read from the amino end (N-terminus) to the carboxyl end (C-terminus). An example of primary structure would be Tyr-Cys-Arg-Phe-Leu-Val-…
Secondary structure (2°) consists of the local three-dimensional structures that form from interactions of the amino acid backbones, like hydrogen bonding. Two common structures that result from these interactions are the alpha helix and the beta-pleated sheet.
Alpha helices are coils occurring from the H-bonds between the N-H and C=O groups along the backbone of the protein.
Beta-pleated sheets are laterally connected strands or sheets of amino acids occurring from the H-bonds between N-H and C=O groups along the backbone of the protein.
Tertiary structure (3°) is the overall 3D structure of the peptide chain. This structure is made by interactions between the R groups of the amino acids.
Quaternary structure (4°) is found in proteins that consist of multiple peptide chains. The individual peptide chains are assembled together to make a multimeric protein structure.
Disruption to Structure
The weak interactions that govern the secondary and tertiary structure of proteins can be disrupted through the application of energy, removal of the water environment, alteration of salt content or changes in the pH. The unfolding or misfolding of proteins from these disruptions is called denaturation. Changes in salt and pH will disrupt the charges on amino acids to disrupt structure. Since eggs are mostly protein and water, it is understood how frying an egg can push the water out (as steam) to leave behind a a congealed gelatinous solid. Similarly, cooking generally works to denature proteins. As such, there are methods of cooking that rely , not only on heat, but on dehydration or curing (using salts) and changes in pH (using acids or bases).
Diversity of Proteins
Proteins are very diverse, both in structure and function. Click on the image below to visit the Protein Data Bank and explore this diversity.
Print this page









