Contents
Virus Exploration
https://media.hhmi.org/biointeractive/click/virus-explorer/
Viruses Reveal the Secrets of Biology
Virus induced cancers and proto-oncogenes
- Read more on the centennial of Rous’ publication.
- Read more on the history of Retroviral oncogenes.
- Read more on the history of oncogene discovery.
Peyton Rous was interested in the transplantability of tumors. In 1910, a rock hen was brought to the Rockefeller Institute with a large tumor. Rous found that a filtrate from the tumor, devoid of cells, had the capacity to induce tumor formation in other chickens in 1911. This led to the idea of an infectious agent that could cause cancers. He would receive the Nobel Prize for this in 1966 due to the subsequent ability of infecting isolate avian cells in culture by Temin and Rubin in 1958. Howard Temin and David Baltimore would later independently discover and characterize an RNA-dependent DNA polymerase we know as Reverse Transcriptase from the Rous Sarcoma Virus that was pivotal for the life cycle of this tumor inducing virus. This resulted in a shared Nobel Prize in 1975.
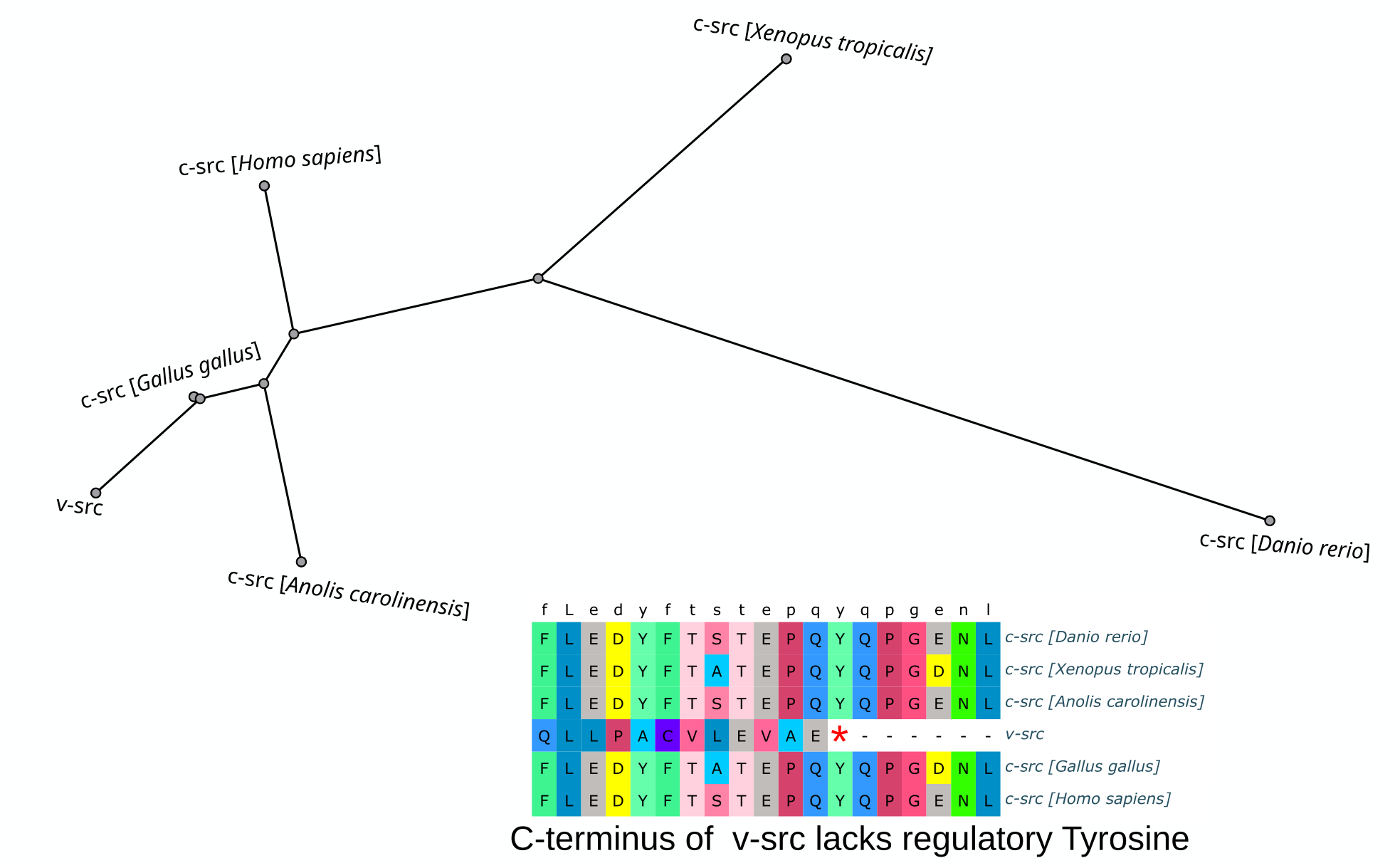
The Nobel Prize was awarded to Michael Bishop and Harold Varmus for illustrating the cellular origin of retroviral oncogenes. This work was initially published in 1976 and demonstrated that v-src in particular, was nearly identical to avian c-src. The main difference in the highly conserved protein was the absence of the C-terminal autoinhibitory Tyrosine residue that resulted in constant activity of the enzyme.
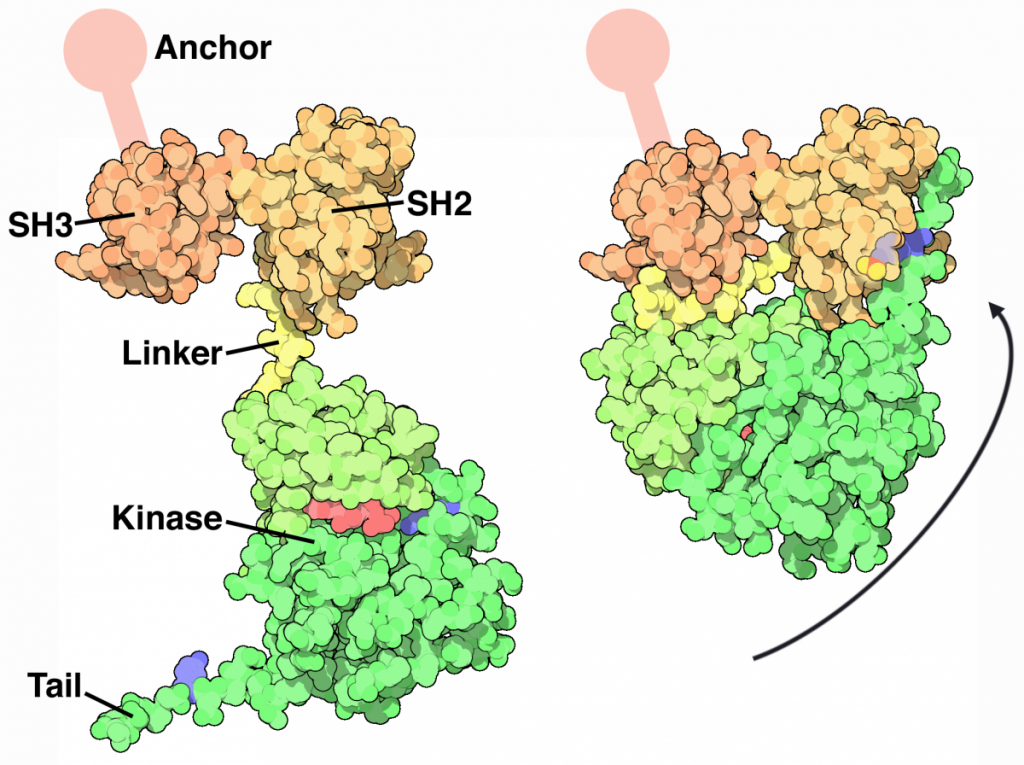
Read more about src
An interview with Harold Varmus from the Journal of Clinical Investigations.
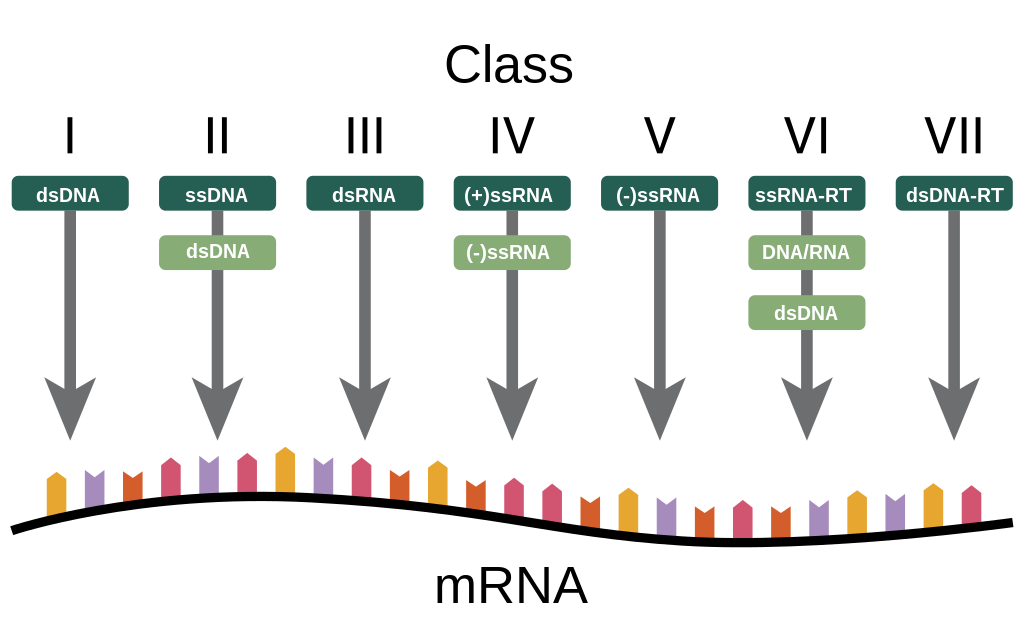
Viral classification
Viral life cycle
Teven Bacteriophage

Genomes of Teven bacteriophages are ~169kb of DNA and code for 300 proteins.
Influenza A
Influenza A is an orthomyxovirus that contains a segmented genome of 8 negative strand RNA molecules encoding at least 10 proteins. Segmentation of the genome increases the the diversity of the virus in rare cases of double infection where each RNA segment acts almost like a chromosome that is packaged in the appropriate complement of segments per functional virion. This shuffling of segments can result in antigenic shift, the shuffling of surface antigens on the surface of the virus particle. The surface antigens are encoded by the hemagglutinin (H) and neuraminidase (N) genes. Hence, the strain of flu viruses are often referred to by the types of these antigens, such as H1N1. This shuffling can also result in hybrid viruses that express the enzymology that has been optimized to specific hosts.
 In addition to the surface antigens, local isolates of influenza are named by their strain type, geographical location, lineage number and year of isolate. Hence, the reconstructed virus from the 1918 pandemic was isolated from a body in permafrost in Brevig Mission, Alaska and is called A/Brevig Mission/1/1918 (H1N1). Learn more about this deadly virus here.
In addition to the surface antigens, local isolates of influenza are named by their strain type, geographical location, lineage number and year of isolate. Hence, the reconstructed virus from the 1918 pandemic was isolated from a body in permafrost in Brevig Mission, Alaska and is called A/Brevig Mission/1/1918 (H1N1). Learn more about this deadly virus here.
-
- Reconstruction of influenza viruses uses cloned segments of the RNA into plasmids. Vaccine strains against specific surface antigens uses the specific H/N combination with the remaining 6 segments that are transfected into susceptible host strain cells where the viruses will multiply and package into infectious particles.
Measles and other Paramyxoviruses


-
- Paramyxovirus genomes are negative strand RNA molecules. The L protein corresponds to the viral polymerase which acts as an RNA-dependent RNA polymerase and replicase. The 5′ leader of this RNA serves as the promoter to continue creating sense strand RNAs for translation.
Flaviviruses
Flaviviruses like Dengue, Zika and West Nile Virus contain positive single-stranded RNA genomes that code for three structural proteins (capsid protein C, membrane protein M, envelope protein E) and seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, NS5).
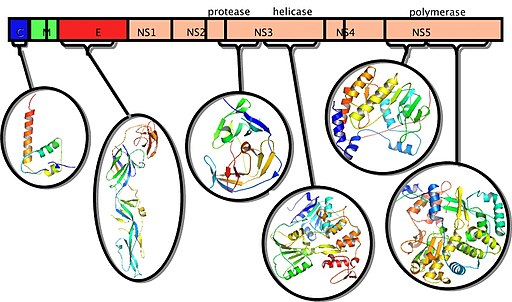



http://pdb101.rcsb.org/motm/103
http://pdb101.rcsb.org/motm/197
Coronaviruses are (+) ss-RNA viruses with genomes ranging from 26.4 to 31.7 kb. two thirds of this genome is occupied by two reading frames called ORF1a and ORF1b. These large open reading frames are translated into large polyproteins that are later cleaved into smaller non-structural proteins. More information on coronavirus proteases involved in these processing steps can be visualized at PDB MOTM.
HIV




 |
 |
 |
 |
Ebola

http://pdb101.rcsb.org/motm/178