tRNA
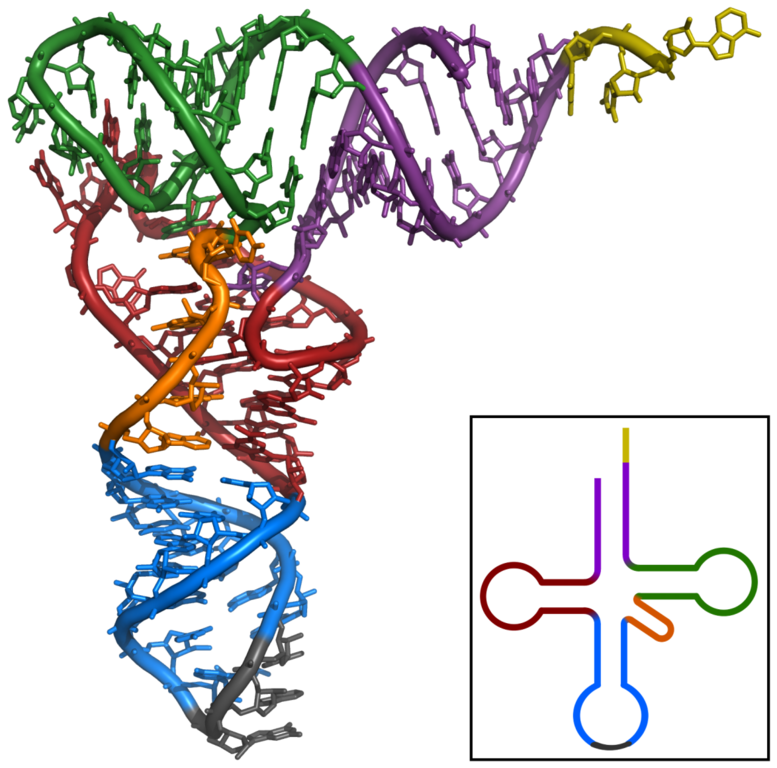
tRNA adapts the RNA and protein world. Each tRNA is unique in the anticodon region that “reads the mRNA. The 3′ end of tRNA, 5′-CCA-3′, is the acceptor stem where a specific amino-acyl transferase attaches the appropriate amino acid to charge the tRNA.

61 types of tRNA may exist within a cell that correspond to the 61 possible coding codons. However, some cells contain less than 61 due to the inclusion of wobble
bases in the anticodon that aid in recognition of redundant codon where the third base is variable with the same amino acid.
Ribosomes
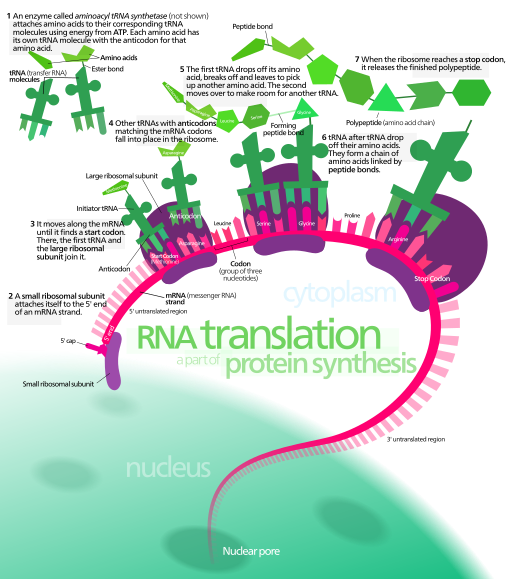
Ribosomes are large complexes of protein and RNA. There are two subunits, a large and small subunit. In eukaryotes, the large subunit is 60S and the small subunit is 40S with an assembled size of 80S. Prokaryotes have a large subunit of 50S and small subunit of 30S with an assembled size of 60S. The small subunit comes in contact with the mRNA and assembles around it.

In prokaryotes, the 16S rRNA contains a small sequence that specifies the reverse complement of the Shine-Dalgarno sequence. mRNA from prokaryotes contain the Shine-Dalgarno sequence that pauses the ribosome to signify that the next proximal AUG sequence is the correct start codon. This sequence is often referred to as the ribosome binding site (RBS).

In eukaryotes, secreted or membrane bound proteins are generated on the rough endoplasmic reticulum. The mRNA encoding secreted proteins contain codons that correspond to a signal sequence that is recognized by the signal recognition particle once it is translated. Once these amino acids come in contact with the signal recognition protein, the ribosome-mRNA complex is ferried to the endoplasmic reticulum where the growing peptide chain is fed through the ER membrane.
Protein Recycling
Proteasome