Contents
Eukaryotic gene expression
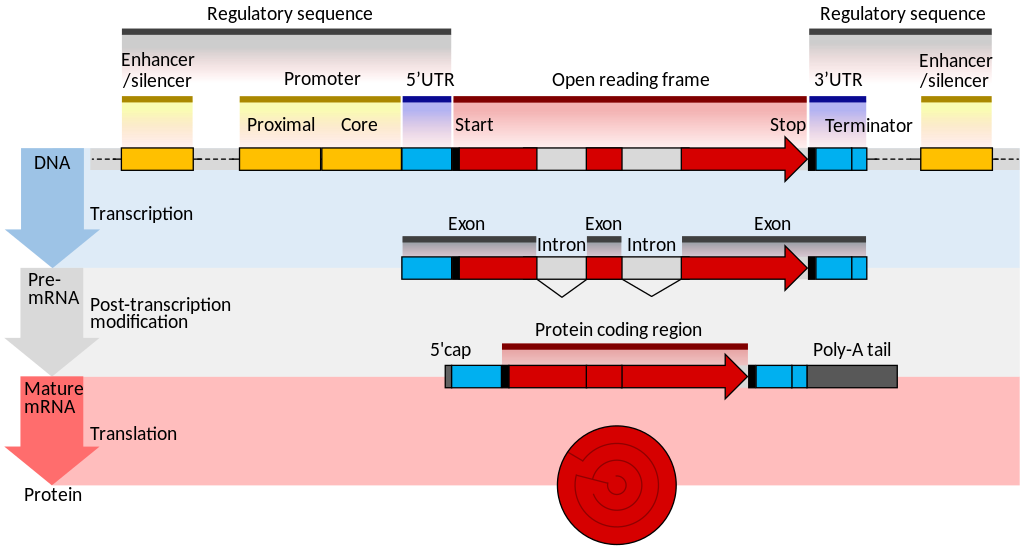
Unlike prokaryotic genes, expression of genes in eukaryotic cells have complex systems of transcription factors that act on promoters to recruit RNA polymerases. Additionally, enhancer elements may reside many kilobase upstream of the promoter. These enhancers strengthen the transcription of the gene. In this case, transcription activator proteins or trans-activators augment the promoter activity.

Transcription Factors
Zinc Finger
The zinc finger domain is generally between 23 and 28 amino acids long containing Cysteine and Histidine residues that interact with zinc ions to form finger-like structures that bind to DNA.
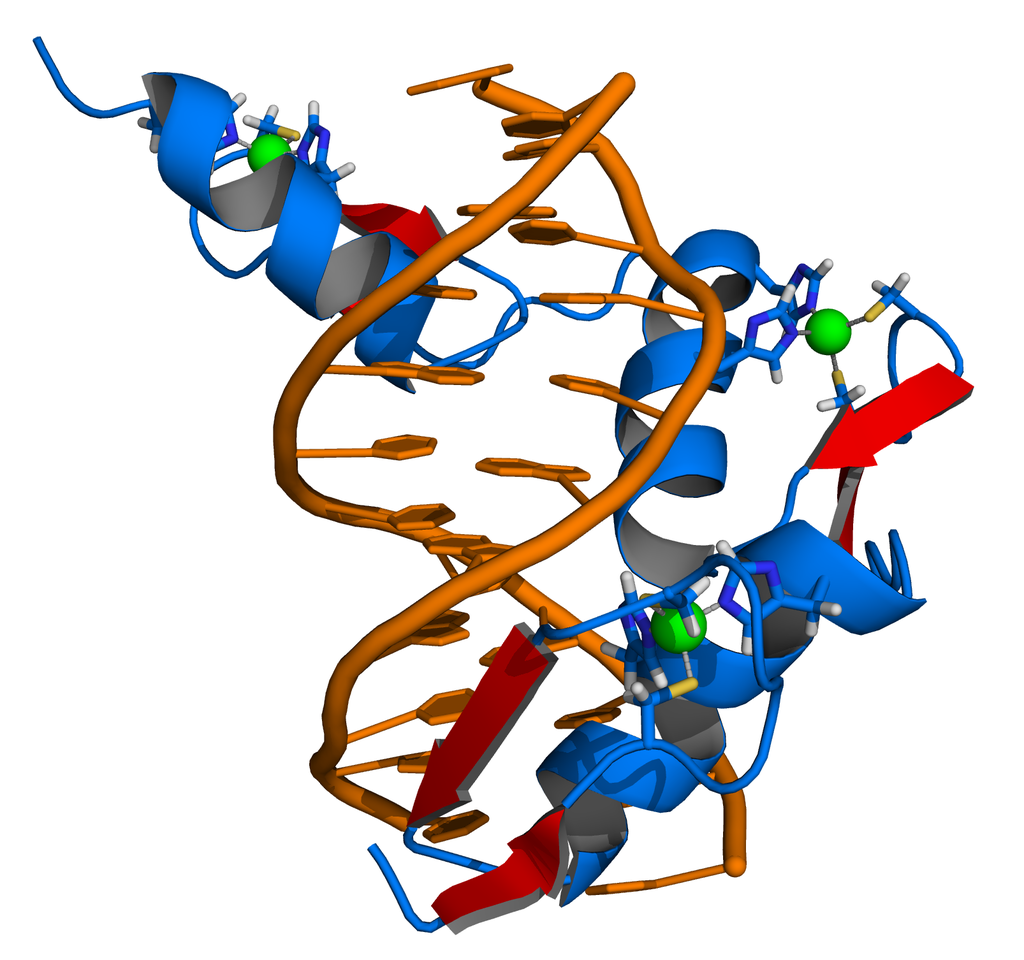
Helix-Turn-Helix
A helix-turn-helix motif contains 2 α-helices containing basic side chains that are spaced apart at the distance of two major grooves.

Leucine Zipper
Leucine zippers are dimeric DNA binding proteins composed of α-helices that cinch two adjacent major grooves. The monomers interacts with each other to form a zipper via amphipathic residues that “zip” together.

Eukaryotic mRNA

Eukaryotic genes may often contain introns (non-coding sequences) that are spliced out from the exons (coding sequences). This complexity permits for increased variety of gene products. Mature eukaryotic mRNAs conatins a 5′-methyl-Guanine followed by an untranslated leader sequence (5′-UTR), the coding sequences (cds), a 3′-untranslated region (3′-UTR) and a long stretch of Adenines (polyA tail).

Expression is most easily measured with RNA since nucleic acid manipulation is fairly simple with 4 different nucleotides. In eukaryotes, the messenger RNA (mRNA) intermediate that is transcribed from DNA contains a polyA tail that is used to separate these messages from other types of RNA that are abundant within cells (like ribosomal RNA). Through the use of an enzyme called reverse transcriptase (RT) and primers composed of deoxy-Thymidines (oligo-dT or dT18), mRNA can be converted into a single strand of DNA that is complimentary to the mRNA. This complimentary DNA is called cDNA. cDNA is very stable compared to the highly labile mRNA and is used for subsequent processing.