Contents
Reading
Chromosomal Basis of Inherited Disease
Learning Objectives
-
- Differentiate between histones and nucleosomes
- Define the terms karyotype
- Describe the differences in organisms with respect to chromsome numbers
- Define the abnormalities in chromosomal numbers: polyploidy, aneuploidy: trisomy and
- monosomy, and mosiacism and their causing mechanisms.
- Define the abnormalities in chromosomal structure: deletions, duplications, translocations, and inversions.
- Distinguish among the following “mistakes” in meiosis: non-disjunction, deletion, duplication, inversion, and translocation; give an example of each.
- Distinguish among Down’s syndrome, Klinefelter syndrome, and Turner syndrome.
- Identify reasons behind epigenetics based on chromosomal structure
- Understand the limitations of Telomeres and Senescence
Chromosomes Overview
Chromosome Packaging

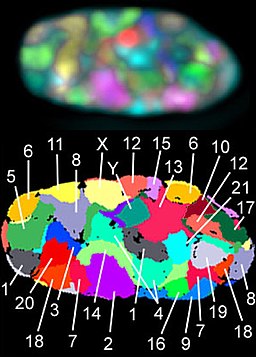
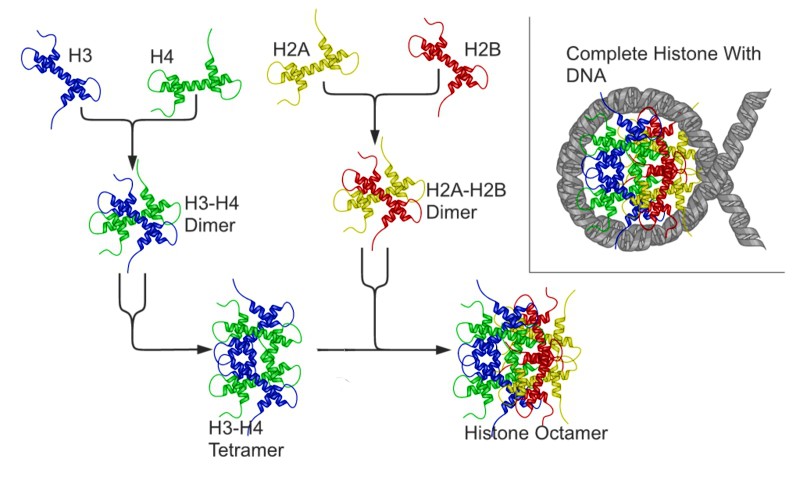
Chromosomes in Interphase are not visible individually and are loosely packaged within the nucleus. In preparation for nuclear division (mitosis or meiosis), they begin to organize tighter and condense in preparation for movement to subsequent daughter nuclei. The animation below illustrates the process of histone packaging and the molecular visualization of DNA replication. Histones are proteins that aid in packaging of the chromosomes into organized coils that give rise to the recognizable chromosomes during metaphase.

DNA is negatively charged due to the Phosphate backbone. Histones are positively charged proteins that associate with eukaryotic DNA to spool and compact the DNA. The form into basic units called nucleosomes. The basic nucleosome core is composed of and octamer of histone proteins (H2A, H2B, H3 and H4).
Chromosome Structure
The familiar X-shape of chromosomes can only be observed during cell division when there are 2 sister chromatids. These chromosomes come in different physical configurations.


Events associated with the improper separation of chromosomes during metaphase results in an alteration of chromosome number in the subsequent generation of cells.