Contents
Lipids Rafts and Membrane microdomains
Signal Transduction
Cell signaling is about the integration of external cues to the inside of the cell to change the physiology of the cell. These signals require the transduction across the plasma membrane. These signals occur via cell membrane receptors that can communicate this information to either act in a rapid manner or in a long-term and slow fashion.

A rapid response is usually mediated via ion channels that open or close in response to an extracellular signal. These are referred to as ionotropic responses.

Metabotropic Receptors
Slower responses enact long-term changes to the cell by activating transcriptional programs and gene expression. These receptors that transduce these signals alter the metabolism of the cell and are called metabotropic receptors.
G-Protein coupled receptors

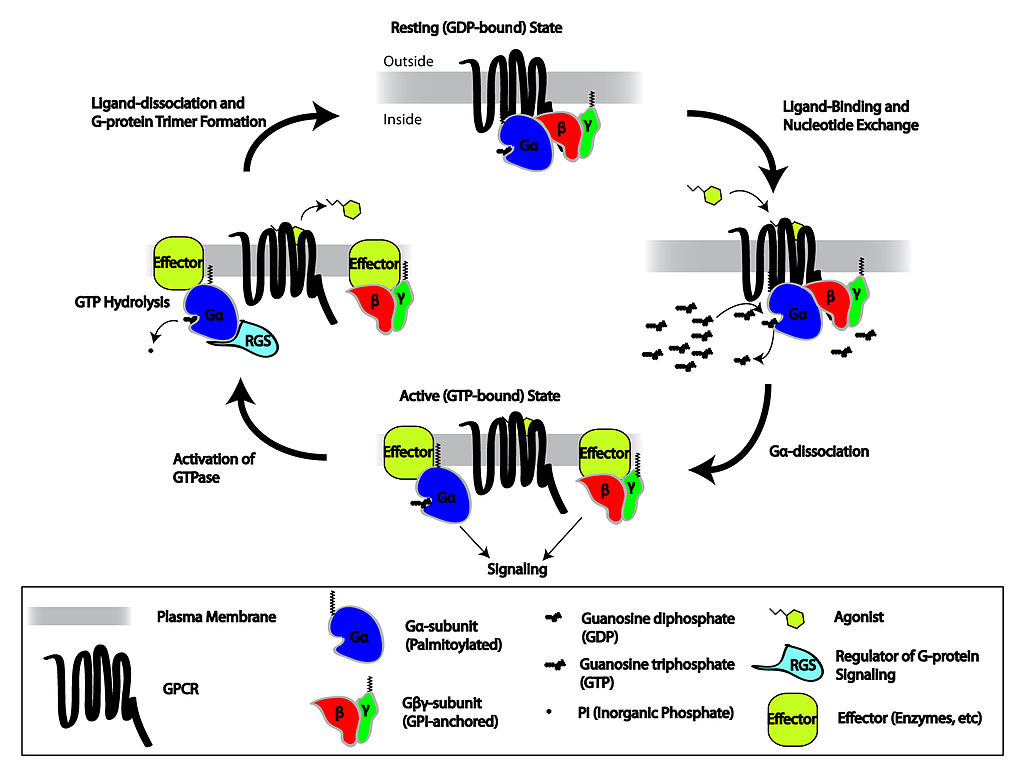
G proteins are GTPases that bind to Guanosine Triphosphate (GTP) or Guanosine Diphosphate (GDP). The GTP bound state will activate an effector protein while the GDP state is inactive. GTPase Activating Proteins (GAPs) activate the enzymatic activity of the G-protein to deactivate the effector. Guanine Echange Factors (GEFs) exchange the GDP with GTP in order to activate the effector associated with the G-protein.

The heterotrimeric G-proteins (consisting of Gα, Gβ and Gγ) are coupled to receptors that aid in direct transduction of receptor binding where the receptor acts as a GEF. Upon ligand activation, Gα becomes bound to GTP and dissociates from the β/γ subunits to activate a downstream effector enzyme. G-protein coupled receptors (GPCRs or 7-transmembrane receptor) represent a common class of metabotropic receptor and most diverse class of proteins represented in the human genome. GPCRs traditionally transduce signals to active adenylyl cyclase (Gαs), inhibit adenylyl cyclase (Gαi)or activate phospholipase C (Gαq).
Protein Kinase A
Gαs activation results in activation of the enzyme adenylyl cyclase to convert ATP into cAMP. Conversely, a GPCR coupled to Gαi inhibits the adenylyl cylcase enzyme.

cAMP, in turn, activates the Protein Kinase A (PKA) serine/threonine kinase. cAMP binds to the regulatory domain of PKA to release the catalytic domain. cAMP is an amplification of the signal being transduced and is referred to as the second messenger. PKA is therefore referred to as the second effector. The signal of cAMP is terminated by a phosphodiesterase (PDE) enzyme that converts it to AMP.

Putative PKA targets can be explored through phosphomimetic or phosphonull mutations in an overexpression system. Since PKA is a serine/threonine kinase, pivotal residues in target proteins can be mutated to an alanine or valine which sterically resemble serine/threonine without the capacity for phosphorylation (phosphonull). Conversely, serine/threonine can be replaced with phosphomimetic mutations by steric and charge analogs like aspartate and glutamate.

Additional reading on cAMP pathway.
Phospholipase activity
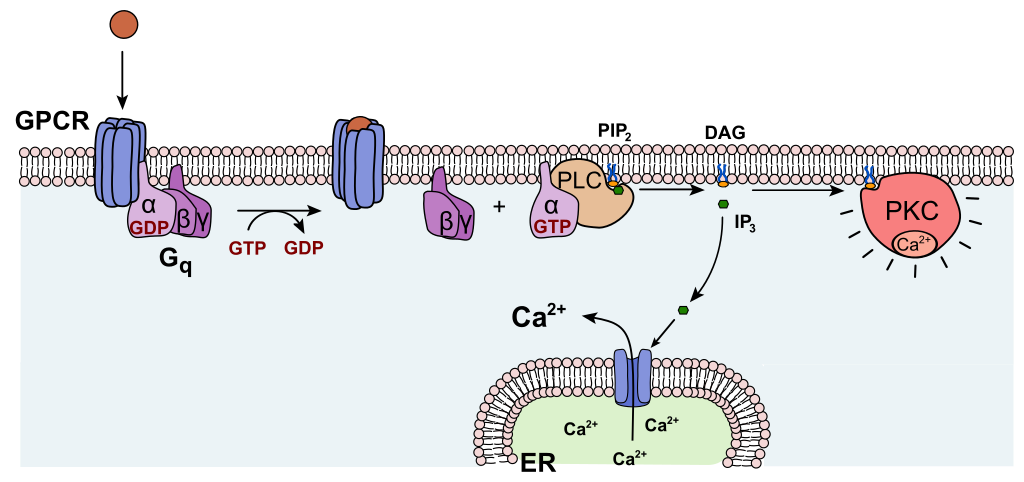
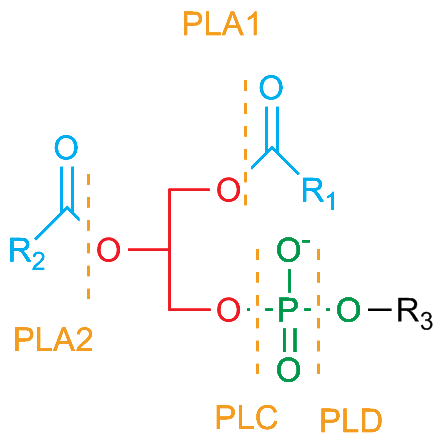
GPCRs coupled to Gαq activate the enzyme Phospholipase C (PLC) which hydrolyzes the membrane lipid Phosphatidyl Inositol bisphosphate (PIP2) to generate a Diacyl Glycerol (DAG) molecule and a inositol triposphate (IP3). DAG unlocks Protein Kinase C (PKC) by displacing a pseudosubstrate domain. IP3 is not tethered to the membrane and activates Ca2+ channels on the endoplasmic reticulum to elevate cytosolic Ca2+. These Ca2+ activate PKC as well as Calcium-Calmodulin kinases. These in turn activate proteins through phosphorylation of serine/threonine residues.

Enzyme-Linked Receptors
Many enzyme-linked receptors couple external signals directly to the activity of a protein tyrosine kinase.

Cell survival pathways will activate receptor tyrosine kinases that activate the PI3K /AKT pathway.

The Insulin Receptor
Insulin binds to the insulin receptor which dimerizes and undergoes a conformational change to activate the protein kinase domains.

NFκB Pathway
The NFκB pathway was first discovered in B-cells as an enhancer of immune functioning. It was named as the nuclear factor kappa-light chain enhancer of B cells. It has since been found to play a variety of roles in immune responsiveness, cancer and synaptic plasticity. Receptor signalling is transduced and activates IκB Kinase(IKK) which in turn phosphorylates the protein IκBα. Under normal circumstances, IκB is complexed with NFκB in the cytoplasm. when IκBα is phosphorylated, it is targeted for degradation by the proteosome and releases NFκB which subsequently dimerizes and enters the nucleus to act as a transcription factor.

Mitogen Activated Protein Kinase
The Mitogen Activated Protein Kinases (MAPKs) are a series of serine/threonine kinases that respond to growth factors. The MAPK pathway is a signalling cascade where MAP Kinase Kinase Kinases (MAP3Ks) activate MAP Kinase Kinases (MAP2Ks) to activate MAPKs. The activation of MAPKs results in the phosphorylation and activation of transcription factors in the activator protein 1 (AP1) family which include: jun, fos and ATF. These proteins dimerize to form leucine zippers and activate transcription at specific promoters.
Mating of Yeast
 Haploid yeast are either a-type or α-type which corresponds to a pheromone produced in order to attract the opposite type. Each yeast type have specific transcriptional programs:
Haploid yeast are either a-type or α-type which corresponds to a pheromone produced in order to attract the opposite type. Each yeast type have specific transcriptional programs:
- a-specific transcriptional program (expressing STE2 /repressing STE3)
- α-specific transcriptional program (expressing STE3/repressing STE2)
STE2 and STE3 correspond to GPCRs that recognize α-factor and a-factor respectively. When a yeast senses the cognate pheromone, the morphology is altered to migrate towards the signal. The yeast are said to adopt a “schmoo” shape during this activation. The haploid yeast will fuse and form a diploid yest.
Read about this at https://www.khanacademy.org/science/biology/cell-signaling/mechanisms-of-cell-signaling/v/overview-of-cell-signaling