- Define: gene, codon, reading frame
- List the major pieces of evidence that led to the hypothesis that DNA influences the production of proteins.
- Define and describe the process of transcription.
- Define introns and exons. Discuss their occurrence and possible roles.
- Summarize the sequence of events that occur in translation.
- Describe the functions of mRNA, tRNA, and rRNA.
- Distinguish between the processes of initiation, elongation, and termination in protein synthesis.
Contents
Genes
Central Dogma of Molecular Biology

Flow of information in cells. DNA serves as a template for copying itself (replication). DNA can also serve as a template for RNA (transcription). RNA is decoded into amino acids to generate proteins (translation). Credit Daniel Horspool (CC-BY-SA 3.0)
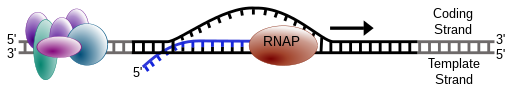
Transcription
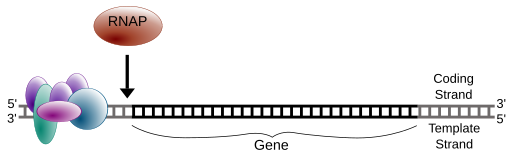
DNA is simply a storage vessel of genetic information. It sits in the nucleus and must be called upon through a process of transcription where an enzyme called RNA Polymerase “reads aloud” the stored information into a molecule called messenger RNA (mRNA). Since DNA is double-stranded in an anti-parallel fashion, we automatically know the sequence of the second strand by knowing the first. The mRNA is made through complimentary base-pairing to the template strand, which is the reverse complement of the coding strand. The coding strand is the strand that reads identical in sequence to the mRNA with the exceptions of T’s being replaced by U’s.
Prokaryotic Transcription
Prokaryotic transcription of genes is driven by the binding the RNA Polymerase subunit called σ factor. A DNA sequence within the promoter for a gene serves as a binding site for σ factor protein which then recruits the rest of the RNA polymerase enzyme to read the DNA template and polymerize messenger RNA. The σ factor binding site determines the directionality of the RNA polymerase, since there is an option of transcribing in 2 directions. The standard σ factor binding site is often denoted by the consensus -35 TTGACA…TATAAT -10 from the transcription initiation. The -10 sequence is sometimes referred to as the Pribnow box.
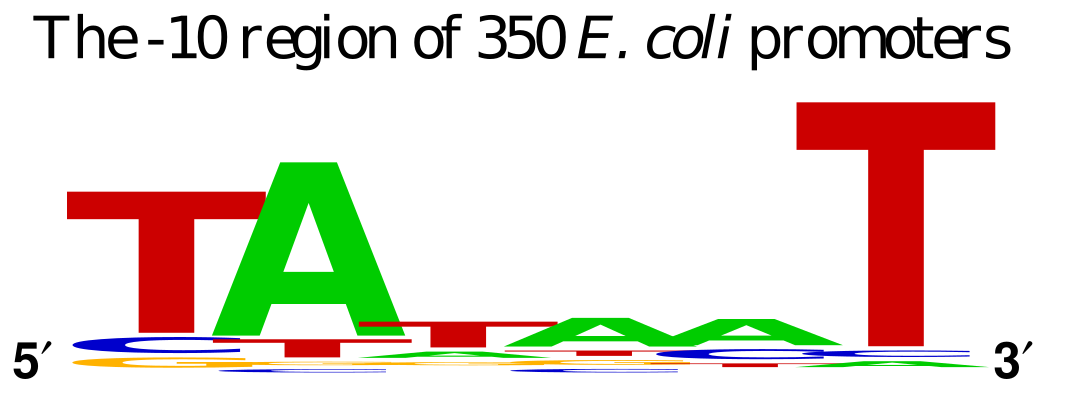
Prokaryotes have a number of σ factor proteins that are involved in response to specific stimuli where the consensus sequence of those promoters is different than the general housekeeping promoters.
Eukaryotic mRNA

Eukaryotic transcription involves more players. Promoter regions of genes in eukaryotes contain a highly conserved region called the TATA box. The name derives from the alternating sequence of T and A nucleotides which are bound to by a protein called TATA Binding Protein (TBP). TBP binding helps to recruit additional transcription factors that then recruit RNA Polymerase II, one of 3 RNA Polymerases, to transcribe the mRNA. RNA Polymerase I is largely responsible for transcription of ribosomal RNAs (rRNAs) while RNA Polymerase III mainly plays a role in transcription of tRNAs.
 General Transcription factors bind to promoters of their respective genes and recruit RNA Pol II. Initiation of transcription occurs in a 5′-3′ direction reading off of the template strand from 3′-5′. The major difference in the structure of prokarytotic genes compared to eukaryotic genes is that eukoryotic genes are often interrupted by non-coding sequences called introns tha must be processed out during the maturation of the nascent pre-mRNA into the mature mRNA that leaves the nucleus for translation.
General Transcription factors bind to promoters of their respective genes and recruit RNA Pol II. Initiation of transcription occurs in a 5′-3′ direction reading off of the template strand from 3′-5′. The major difference in the structure of prokarytotic genes compared to eukaryotic genes is that eukoryotic genes are often interrupted by non-coding sequences called introns tha must be processed out during the maturation of the nascent pre-mRNA into the mature mRNA that leaves the nucleus for translation.
Eukaryotic RNA Processing

Eukaryotic pre-mRNA undergoes processing to mature into an mRNA ready for translation. These modifciations include intron splicing, poly-Adenylation and addition of a 5′-cap.

Schematic diagram of the 5′-mG cap composed of 7-methylguanosine. This modified base is found at the 5′ end of every mature mRNA in eukaryotes and aids in translation.
Eukaryotic genes may often contain introns (non-coding sequences) that are spliced out from the exons (coding sequences). This complexity permits for increased variety of gene products. Mature eukaryotic mRNAs conatins a 5′-methyl-Guanine (mG-cap) followed by an untranslated leader sequence (5′-UTR), the coding sequences (cds), a 3′-untranslated region (3′-UTR) and a long stretch of Adenines (polyA tail).
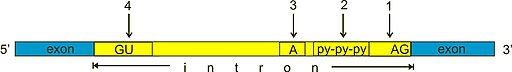
Schematic diagram of the exon/intron boundaries as well as consensus sequences found at the edge of introns.

Most Eukaryotic genes undergo splicing which is carried out in a series of reactions catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). Sometimes, these introns self-splice due to inherent enzymatic activity of the RNA. These catalytic RNAs are termed ribozymes.
 Introns are spliced out of the mature mRNA but may also involve alternative splicing where exons are also excised from the sequence. This exon skipping ultimately produces multiple protein products from a single gene that can diversify the activity or localization of the protein within the cell.
Introns are spliced out of the mature mRNA but may also involve alternative splicing where exons are also excised from the sequence. This exon skipping ultimately produces multiple protein products from a single gene that can diversify the activity or localization of the protein within the cell.
Expression is most easily measured with RNA since nucleic acid manipulation is fairly simple with 4 different nucleotides. In eukaryotes, the messenger RNA (mRNA) intermediate that is transcribed from DNA contains a polyA tail that is used to separate these messages from other types of RNA that are abundant within cells (like ribosomal RNA). Through the use of an enzyme called reverse transcriptase (RT) and primers composed of deoxy-Thymidines (oligo-dT or dT18), mRNA can be converted into a single strand of DNA that is complimentary to the mRNA. This complimentary DNA is called cDNA. cDNA is very stable compared to the highly labile mRNA and is used for subsequent processing. While all life on this planet share a common molecular toolbox, the presence of introns in a eukaryotic gene prohibits direct expression within a prokaryotic system. In the case of expressing a eukaryotic gene in a prokaryote, the cDNA version of that gene would be used within a prokaryote since the introns have been processed already.