Contents
Cytogenetics and Karyotyping
Objective
This week we will be learning how cytogeneticists use karyotyping to understand chromosomal abnormalities. Karyotyping is a technique where chromosomes are stained and visualized during the metaphase stage of cell division (mitosis or meiosis). In this lab, you will take on the role of a medical cytogeneticist and use human karyotypes to diagnose various diseases and abnormalities in patients. Upon completion of the lab, you should be familiar with what chromosomes are, what a karyotype is and how it is constructed, the different ways in which chromosomal abnormalities might arise, and the basic terminology used to describe chromosomes.
Background information
Overview
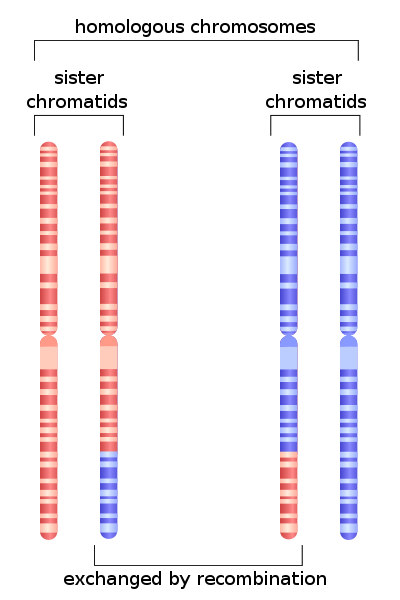
DNA is packed into units called chromosomes. In many species, DNA is closely associated with several types of proteins called histones that are used to tightly pack the DNA into the cell nucleus (in eukaryotes). In eukaryotes, we say that the chromosomes are linear, whereas most bacteria contain a single circular chromosome. All eukaryotes also contain a second genome inside their cells called mitochondrial DNA (mtDNA). Genes in mtDNA help regulate the process of cellular respiration. In addition, plants and several protists contain a third genome in their chloroplasts called chloroplast DNA (cpDNA), which is involved with photosynthesis. In this lab we will focus exclusively on eukaryotic nuclear chromosomes. Human somatic cells contain 46 chromosomes. As humans are a diploid species, half of the complement of chromosomes (23) originated from a sperm cell and half (23) originated from the egg cell. These are called homologous chromosomes. For example, each person has two copies of Chromosome 1, one copy inherited from the father and one copy inherited from the mother. Portions of homologous chromosomes can exchange segments during meiosis in a process called recombination. During cell division, each homolog is also duplicated, forming sister chromatids (Fig. 1). The region where sister chromatids attach during cell division is called the centromere. Telomeres are regions at the tips of chromosomes that consist of highly repetitive sequences that form a protective cap to the ends of chromosomes. Telomeres tend to shorten with each cell division, leading to cell aging and eventually cell death. Much research has focused on techniques to decrease the propensity of telomere shorting to counteract the aging process. Therapeutic-related work has also focused on the role of telomeres in the proliferation of cancer cells.
Human sex is determined genetically through the XY system—human females are XX and human males are XY. Research has shown that human “maleness” is determined by a gene that sits on the Y chromosome. In several other species, sex is not determined by genes and chromosomes, but by environmental temperature though what is called temperature-dependent sex determination. For example, in many crocodilian species, males are only produced if eggs are incubated at intermediate temperatures. In contract, females are produced if eggs are incubated at either extreme.
There is a large variation in the number of chromosomes among species, but there tends to be a correlation between organismal complexity and genome size. The human genome is approximately 3 Gbp, or 3 billion base pairs long. This is considered a relatively large genome (some salamander genomes are much larger), but surprisingly only about 2% of the genome encodes a functional product (e.g. proteins). Many researchers refer to the remaining 98% as ‘junk DNA’, although it is likely that other regions of the genome are involved in regulating gene expression to some degree. For such a large genome to fit inside the cell nucleus, it needs to be condensed substantially. The DNA is first wrapped around histone proteins, which then associate to form what are called nucleosomes. The nucleosomes further compress into what is called a 30 nm fiber. Additional coiling eventually gives chromosomes their highly compact, typical appearance during cell division.
Chromosome classification
Chromosomes are generally classified using multiple criteria. First, they are number from largest to smallest. For example, human chromosome 1 would be the largest chromosome, which contains 2,100 protein-coding genes and 249 million bp. The sex chromosomes are labeled appropriately as either XX in females or XY in males. Note that the X-chromosome is much larger than the Y-chromosome. Therefore, the X- and Y- chromosomes are considered non-homologous (although there are a few homologous regions that are needed for proper pairing during cell division).
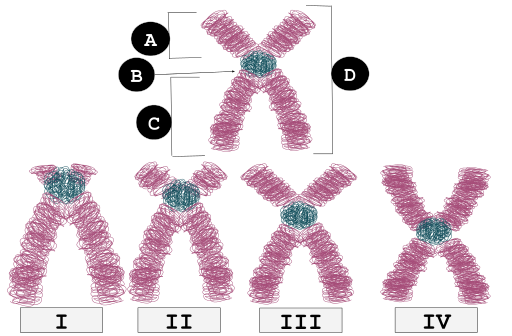
The relative position of the centromere can also differ between chromosomes. Centromeres that are placed in the center of a chromosome are called metacentric chromosomes, resulting in equal length chromosomal arms. When the centromere is not found in a central position, different chromosomal arm lengths result. These arms are referred to as p arms (short arms) and q arms (long arms). In addition to metacentric chromosomes, chromosomes can be submetacentric, acrocentric, and telocentric, all varying in the relative position of the centromere (Fig. 2).
A final way that cytogeneticists can differentiate and classify chromosomes is to stain them with dyes that produce diagnostic banding patterns. Note that bands on a stained chromosome are not indicative of genes. In general, there can be dozens or hundreds of genes within a single band. A common stain used by cytogeneticists is called the Giemsa stain, also known as the G-banding technique. Following the staining procedure, AT-rich regions of the chromosome appear dark whereas transcriptionally active GC-rich regions incorporate less of the dye and thus appear lighter. The differential banding patterns can be used to identify homologous chromosomes and look for chromosomal alterations.
Procedure for creating karyotype
- Obtain cells (e.g. white blood cells, skin cells, cancer cells, cells from amniotic fluid). Obtaining cells from amniotic fluid (amniocentesis) can be used to detect genetic abnormalities in the fetus.
- Treat cells with chemical to promote cell division.
- Treat cells with a second chemical that halts cell division at metaphase. This is the stage when chromosomes are highly condensed and easily visualized.
- Immerse cells in a hypotonic solution and place on glass slide.
- Stain chromosomes with Giemsa stain and observe under microscope.
- Prepare digital images and analyze results.
Genetic abnormalities observed by G-banded karyotyping
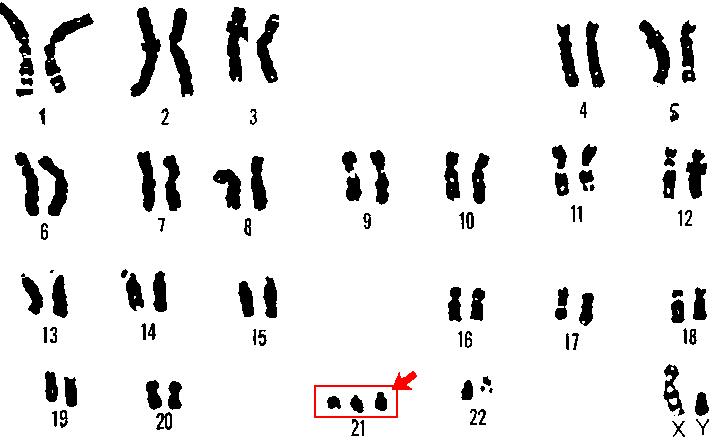
In general, chromosome abnormalities can include both numerical changes and structural changes. Numerical changes can involve an irregular number of a particular chromosome (aneuploidy). For example, a diploid individual possessing three copies of a chromosome instead of the usual two copies would be called trisomy, whereas an individual with one copy would be called monosomy. Trisomy 21 would mean three copies of chromosome 21 and is characteristic of individuals with Down syndrome (Fig. 3).
In addition to abnormalities in the number of specific chromosomes, an individual can have an incorrect number of the complete set of chromosomes. A gain in one or more complete sets of chromosomes is termed euploidy, whereas a loss of an entire set of chromosomes is called monoploidy. Monoploidy is usually fatal due to recessive mutations (many of which are lethal) being phenotypically expressed.
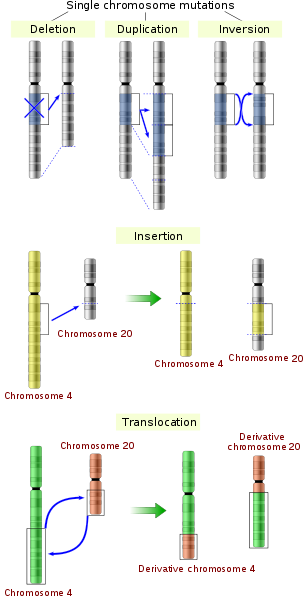
In addition to numerical changes, chromosomes can also exhibit the following structural changes involving either one or two chromosomes (Fig. 4):
Translocations (two chromosomal change) – two non-homologous chromosomes fuse or swap segments.
Insertions (two chromosomal change) – one region of one chromosome removed and inserted into a different chromosome.
Inversions (single chromosomal change) – certain regions of a chromosome are placed in an incorrect orientation with respect to the remainder of the chromosome.
Duplications (single chromosomal change) – one segment of a chromosome is duplicated.
Deletions (single chromosomal change) – removal or loss of a particular region or segment of a chromosome. Commonly occurs in telomeric regions during cell division.
Although standard karyotyping can provide a powerful approach to diagnosing various genetic diseases, it is considered a wide resolution technique, meaning that it is unable to detect genetic abnormalities below a certain number of base pairs (~1 million). In these cases, other techniques such as fluorescent in situ hybridization (FISH), array comparative genome hybridization (CGH), and direct DNA sequencing can be used, the latter of which can be used to detect single nucleotide changes or polymorphisms (SNPs).
Activity
Working in pairs, you will assume the role of a medical cytogeneticist and aid in the diagnosis of various genetic diseases. Each group should try to get through as many case studies as possible.
- Each group should receive several blank copies of the Cytogenetics Report to be filled out for each case study.
- Each Chromoscan Board contains a single case study, is color-coded and is associated with the colored chromosome decals. For example, Case Study A is pinkish purple. Never mix and match Chromoscan Boards and decals.
- Randomly select one of the chromosome decals and make a sketch of the chromosome on the Cytogenetics Report. Be sure to label the centromere, telomere, p arm, and q arm. Also make note if the chromosome is metacentric, submetacentric, acrocentric, or telocentric.
- Record all the relevant information from the case study on the report.
- To create the karyotype you will match the correctly colored chromosome decals with the chromosomes already present on the Chromoscan Board. In other words, your goal is to correctly pair homologous chromosomes.
- Once the karyotype is created, analyze it for any chromosomal abnormalities (e.g. inversions, translocations, deletions, insertions, duplications).
- Report your findings on the Cytogenetics Report.
- To diagnose the patient, refer to the table below and write the diagnosis on the Cytogenetics Report.
- Using your smartphones, provide additional clinical information (in addition to what is presented in the case study) about the disease on the Cytogenetics Report.
- Remove chromosome decals and randomly place them in the cryostorage region of the Chromoscan Board.
- Repeat procedure for additional case studies. Do not share answers with other groups!
Diagnostic Table
| Karyotype Findings | Diagnosis |
| No abnormalities found | None |
| Trisomy 21 | Down syndrome |
| XXX | Trisomy X female |
| Inversion on chromosome 3 involving p arm, centromere and q arm | Not associated with any known disease |
| Trisomy 18 | Edwards syndrome |
| Trisomy 13 | Patau syndrome |
| Reciprocal translocation with chromosomes 9/22 | Chronic myelogenous leukemia |
| Robertsonian translocation (fusion of chromosome arms) with chromosomes 14/21 | Down syndrome |
| XXY | Klinefelter syndrome |
| XYY | XYY male |
| 5p deletion | cri du chat |
| Monosomy X | Turner syndrome |
| Inversion on chromosome 9 | Not associated with any known disease |
Cytogenetics Report
Names:_______________________________________ Date______________
Directions: Each group should complete a separate Cytogenetics Report for each case study (print out several sheets). After a case study is completed, place the chromosome decals randomly in the cryostorage regions of each board. Do not mix and match chromosomes and boards. Before assembling the karyotype, randomly select one chromosome and make a detailed sketch in the space below. Note the location of the centromere, telomeres, the p arm, and the q arm. Also note if the chromosome is metacentric, submetacentric, acrocentric or telocentric.
Chr. type: ______metacentric _____submetacentric _____acrocentric _____telocentric
| Patient name | Case Study ID | Patient Age |
| Why is the patient being referred for karyotyping? | Source of cells:
______Blood ______Amniocytes ______Chorionic Villi ______Other |
|
| Total number of chromosomes observed | Gender | |
| Chromosomal Findings
_____no abnormalities _____monosomy (chr____) _____trisomy (chr ____) _____deletion (chr ____, arm _____) _____insertion (chr_____, arm_____) _____translocation (chrs____and____) _____inversion (chr____, arm____) |
Patient diagnosis and additional information to provide to patient and family. | |
Preparation of materials for Lab 3
Your final objective this week is to prepare fungal genetic crosses to observe the following week. This experiment provides a simple, yet effective hands-on introduction to basic properties of meiosis. Our technicians have already prepared cultures of three types of Sordaria fimicola (wild-type, mutant tan, and mutant gray). In the lab you should see six dishes, two containing the wild-type strain, two with the tan strain, and two with the gray strain. Your job is to set-up crosses of these different strains to determine the proportion of asci containing “hybrid” ascospores that are a result of crossing over during prophase I of meiosis. You will have multiple dishes containing agar that will be used to perform your experiment.
- Use alcohol to disinfect all work surfaces and wash your hands.
- Obtain a petri dish containing the Sordaria crossing agar to make your crosses. Remember that we are making crosses between wild-type and gray and wild-type and tan strains. For simplicity, I recommend using separate plates for the tan and gray mutants. For example, one dish will be a wild-type vs. tan cross and another dish will contain wild-type vs. gray cross.
- Using a marking pencil, invert your dish and draw four equal quadrants. Label two of the quadrants as wild-type and two as mutant (either gray or tan). Refer to Fig. 5 for an example. Also, make sure to write your names and the date on your dishes.
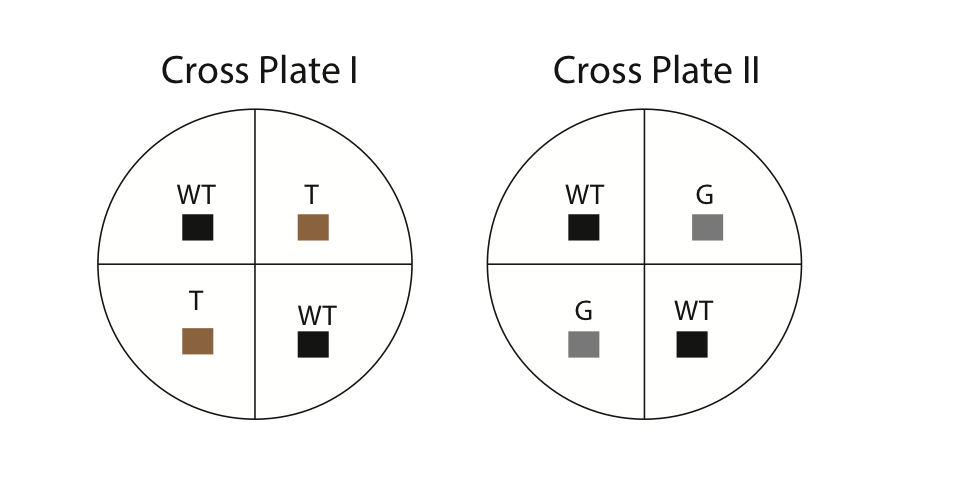
- Using a sterilized wooden splint, cut small 0.5 cm cubes from the stock culture dishes. It is important to use sterilized equipment to minimize contamination. The common procedure for sterilization is to dip a utensil (e.g. scalpel) into alcohol and then flame it over a Bunsen burner. In our case, the wooden splits are already sterilized. Carefully invert the cubes (hyphae side down) and place them in the correct location on the cross plates (see diagram above). For example, you will cut out a 0.5 cm cube from the tan strain and place it in one of the quadrants labeled “tan” on your cross plate. Each culture plate should contain four cubes (two wild-type and two mutant [either tan or gray, but not both]). Using the side of your splint, gently press the cubes onto the agar so they stick. Next, apply some parafilm and seal the outside of your dishes.
- Incubate the dishes at room temperature for 1 week.




