Using the primer sequences, one can determine the size and/or location of a PCR product. This can be done using BLAST or with a program like UGENE.
Using BLAST
- Primers for the PV92 insertion
- Forward primer: 5′ GGATCTCAGGGTGGGTGGCAATGCT 3′
- Reverse primer: 5′ GAAAGGCAAGCTACCAGAAGCCCCAA 3′
- Visit BLAST: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch
- Paste both primers:
- GGATCTCAGGGTGGGTGGCAATGCTGAAAGGCAAGCTACCAGAAGCCCCAA
- Remove the 5′ and 3′ numbers
- Choose “Somewhat Similar“
- Locate the locus of the product and the size
Using Ugene
- Exercise using the human D-Loop Primers
- Forward Primer 5’-TTAACTCCACCATTAGCACC-3’
- Reverse Primer 5’-GAGGATGGTGGTCAAGGGAC-3’
- Download the sample Genbank file: Human Mitochondrial Genome
- Open the file in Ugene
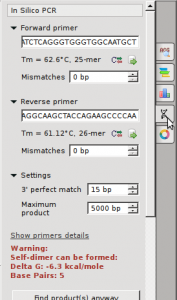
- Select the “In Silico PCR” button on the far right (double helix button)
- insert forward and reverse primers in appropriate spaces
- A PCR product should be noted for one of the sequences after pressing “Find Products anyway“