Contents
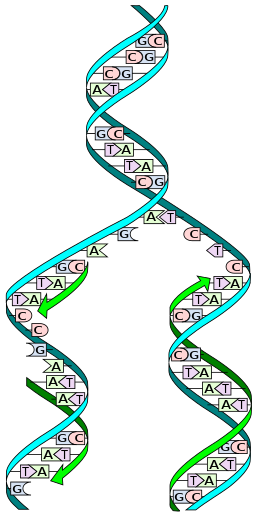
DNA Replication

Meselson & Stahl Experiment

Reference:
Meselson M, Stahl FW. THE REPLICATION OF DNA IN ESCHERICHIA COLI. Proc Natl Acad Sci U S A. 1958 Jul 15;44(7):671-82. doi: 10.1073/pnas.44.7.671. PMID: 16590258; PMCID: PMC528642.
Replication Machinery

Reference:
Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci U S A. 1968 Feb;59(2):598-605. doi: 10.1073/pnas.59.2.598. PMID: 4967086; PMCID: PMC224714.